A major investment by the Government of Canada's Genomics Research and Development Initiative (GRDI) is delivering the science needed to strengthen Canada's ability to address antimicrobial resistance—AMR—widely regarded as one of the most serious health threats facing the world today.
As more and more bacteria and other pathogens such as viruses, fungi and parasites develop resistance to an ever‑wider range of antibiotics and antimicrobials, infections long considered little more than a nuisance are becoming difficult and sometimes impossible to treat. If action is not taken to address the problem, it is estimated that by 2050, AMR could be responsible for as many as 10 million deaths a year around the world.Footnote1
Government-wide collaboration
"To understand whether and how antibiotic use in food production is contributing to AMR in humans, we need to look at and compare bacteria collected from sick people in hospitals, from animals used in food production, and from the environment around beef, pork and poultry operations."
Launched in 2016, the 5‑year GRDI‑AMR project involves a number of interlocking research efforts underway at 5 federal departments and agencies—the Canadian Food Inspection Agency (CFIA), the Public Health Agency of Canada (PHAC), Health Canada (HC), the National Research Council (NRC), and Agriculture and Agri‑Food Canada (AAFC). In all, some two dozen federal government scientists are involved in the research, supported by a host of post‑doctoral researchers and graduate students. The GRDI‑AMR project is the only specifically targeted research investment under the Federal Action Plan on Antimicrobial Resistance and Use in Canada.
One Health
Antimicrobial or antibiotic?
While the terms are often used interchangeably, an antibiotic is an antimicrobial used to treat bacterial infections. Antimicrobials also include substances used to address non‑bacterial pathogens, such as fungi or viruses.
Project coordinator Dr. Ed Topp, a Senior Research Scientist at AAFC, says this broad‑based collaboration is essential. "Bacteria are found in people, in animals and in the environment, and they circulate easily from one to the other—that's why we need the 'One Health' approach. To understand whether and how antibiotic use in food production is contributing to AMR in humans, we need to look at and compare bacteria collected from sick people in hospitals, from animals used in food production, and from the environment around beef, pork and poultry operations."
Keys to progress: genomics technologies and collaboration
With access to state‑of‑the‑art genomics technologies such as whole genome sequencing made possible by the GRDI, researchers can quickly examine the entire genome of a bacteria, and see if it contains any of a range of genes known to be associated with AMR. By comparing the AMR genes in bacteria collected in different sectors, researchers can see, for example, whether the genes that give AMR to a bacteria found in poultry are the same genes associated with AMR in similar bacteria found in humans.
"Thanks to the collaboration enabled by the GRDI, we're able to share the data being generated by the various projects and put it to work in different ways," says Dr. Topp. "At the PHAC, for example, access to all this data is giving Dr. Richard Reid‑Smith and his team the information they need to assemble what will be Canada's first‑ever national AMR risk assessment map—that's a major step toward enabling policy makers and other stakeholders to identify where action on AMR can be most effective. At the CFIA meanwhile, Dr. Catherine Carrillo and her team have been able to add detection of AMR to surveillance of priority pathogens found in food, such as disease‑causing types of E. coli and Salmonella."
Significant findings
"We're also seeing good news in developing alternatives to antibiotics in food production… compounds derived from cranberries and other plants could replace some of the antibiotics used in the poultry industry."
Dr. Topp says GRDI‑AMR research is delivering important results in other ways. "At AAFC in Lethbridge, a team led by Dr. Tim McAllister has been working on connecting antibiotic use in beef cattle to AMR in people—and have not found any evidence of a link."
"We're also seeing good news in developing alternatives to antibiotics in food production," says Dr. Topp. "Here at AAFC for example, Dr. Moussa Diarra and his team are showing how compounds derived from cranberries and other plants could replace some of the antibiotics used in the poultry industry."
Showcasing Canadian innovation
In addition to supporting the development of new strategies to address AMR in Canada, the GRDI‑AMR project is helping to strengthen Canada's global reputation for innovation.
Dr. Gerry Wright, co‑founder of the Comprehensive Antibiotic Resistance Database (CARD), housed at McMaster University in Hamilton, says the GRDI‑AMR project is providing a tremendous amount of new information to the database, which acts as a global hub for information about the genes associated with AMR. "The GRDI researchers are sequencing the genomes of large numbers of bacteria found in the environment, in animals and in humans, giving us—among other things—a new understanding of how the genes associated with AMR circulate," says Dr. Wright. "They are also finding AMR genes that have not previously been identified. Our database is accessed by researchers from around the world on a daily basis, so the GRDI‑AMR project is making a significant contribution to the global effort to address AMR—and that's important, because this really is a global issue."
The GRDI‑AMR project is raising Canada's profile in other ways. Scientists working on the project have been asked to discuss Canada's approach and present their findings at a number of international conferences on AMR, as well as institutions ranging from the World Heath Organization to the U.S. Presidential Advisory Council on Combatting Antibiotic‑Resistant Bacteria.
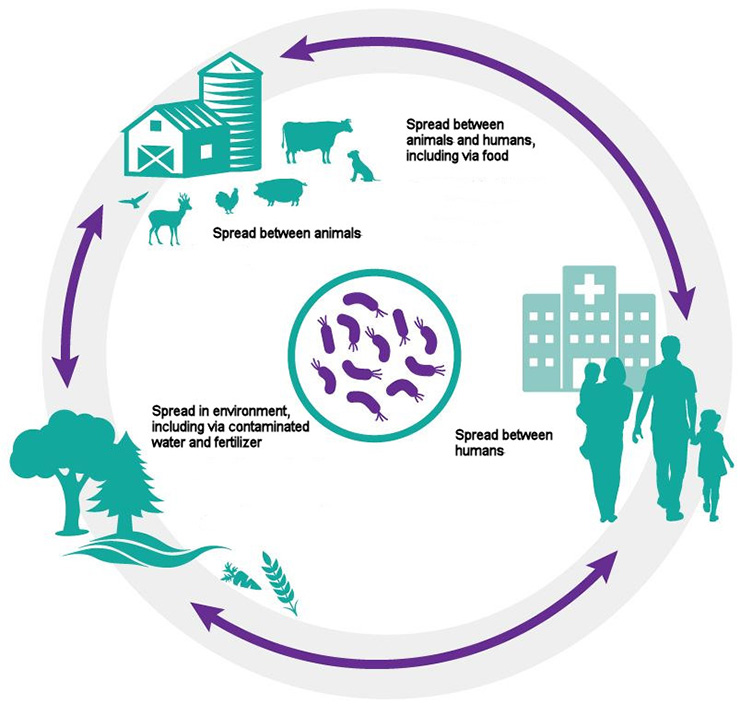
One Health: Bacteria are found in people, in animals and in the environment, and they circulate easily from one to the other.
-
Long description of diagram
Circular diagram showing how bacteria can spread and circulate:
- between animals
- between animals and humans, including via food
- between humans
- in the environment, including via contaminated water and fertilizer