Alertnate format: Genomics R&D Initiative - Science with Impact (PDF, 2.2 MB)
There can be no doubt that research projects funded through the Government of Canada's Genomics Research and Development Initiative (GRDI) are already delivering significant benefits to Canadians and advancing key Government of Canada outcomes—healthy Canadians; strong economic growth; an innovative and knowledge-based economy; a clean and healthy environment.
Impactful science for Canadians
Benefits will increase
"... GRDI-funded projects are likely to have real and lasting longer-term impacts... billions of dollars in benefits for end-users... "
Indeed, in completing a horizontal evaluation of the GRDI in 2016, a leading Canadian management consulting firm predicted that, "GRDI-funded projects are likely to have real and lasting longer-term impacts... billions of dollars in benefits for end users... improved public health and wellness; avoided health system costs; efficiencies and avoided losses for the public and private sectors; environmental sustainability; improved detection of invasive species; improved strains/traits of plants, trees and animals of commercial value; and improved fisheries tracking and management."Footnote 1
Building Canada's global brand

Photo credit: National Research Council of Canada
The achievements recorded to date by GRDI researchers are also winning international recognition for Canada. GRDI research discoveries have earned Canada a place on international bodies developing guidance for the application of genomics in fields ranging from regulatory policy to food safety. GRDI researchers have been praised for their contributions to the global fight to eradicate HIV. Genomics applications developed by GRDI researchers are now in use by agencies in other countries, including the US Department of Agriculture. Literally hundreds of papers by GRDI researchers have been accepted for publication in peer-reviewed journals.
Collaboration across government
Beyond its scientific achievement, the GRDI is also drawing attention for the unique, multi-department collaboration that has demonstrated the value that can be achieved in addressing shared priorities through the coordinated application of complementary expertise and infrastructure from several departments and agencies.
"... at a cost many times less than what other countries have invested in this type of research."
The five-year Food and Water Safety project, for example, involved more than 50 scientific and technical personnel from six federal departments and agencies (DAs). As one senior researcher has said, when presenting the results of the research at international conferences, "There's great interest in what we've been able to achieve—but I've also been asked time-and-again how we were able to bring together such a large team from different disciplines and different departments and get them all working in a collaborative way toward these shared priorities, and at a cost many times less than what other countries have invested in this type of research."
Leveraging investment and expertise
While Government of Canada funding for the GRDI has remained at $19.9 million per year since 2002, GRDI research projects continue to attract important financial and in-kind contributions from other sources—a total of $42.8 million in the 2016-17 fiscal year alone.Footnote 2
Recognizing the importance of genomics research to the execution of their mandates, the bulk of these contributions have come from GRDI-funded departments and agencies, although projects have also leveraged support from granting agencies (Natural Sciences and Engineering Research Council of Canada, Canadian Institutes of Health Research, the Canadian Foundation for Innovation), as well as stakeholders, clients, First Nations, and community groups.
"... GRDI research projects continue to attract important financial and in-kind contributions from other sources... "
The impact of Government of Canada investment in the GRDI is further multiplied by extensive collaboration with researchers outside the federal government—in universities, in particular with scientists supported by Genome Canada and its regional centres, as well as public and private sector entities in Canada, the US, Spain, Germany, the UK and elsewhere. In 2016-17, for example, some 480 external collaborators contributed expertise and financial and/or in-kind resources to GRDI research projects.
The bottom line
Perhaps most important to understanding the positive impact of the GRDI is its fundamental role in supporting mandate-driven genomics research and development—generating results that have been neither pursued nor achieved elsewhere. The GRDI's strategic alignment with federal government and departmental objectives and priorities—its focus on addressing issues of importance to Canadians—makes the GRDI an essential and unique response to a specific need that is not being addressed by other genomics R&D being conducted in Canada.
About this document
Given the wide range of achievement and the extent of the progress GRDI-funded research has delivered in areas that matter most to Canadians, it is impossible to detail every aspect of the real and potential impact of the Government of Canada's investment in the GRDI in this brief report; rather, primarily through the use of selected examples and discussion of the management and governance of the GRDI, it is intended to provide the reader with a degree of insight into the true significance of the initiative.
A model of interdepartmental collaboration
"The introduction of interdepartmental SPPs in Phase V [beginning in 2011] has proven to be a strong feature of the program... The two SPPs funded in Phase V were found to be well-managed and have achieved significant results, which likely would not have occurred in the absence of GRDI funding for the interdepartmental nature of the projects."
The desirability of greater interdepartmental cooperation—often referred to as 'breaking out of silos'—has been a recurring policy theme within the Government of Canada for some time. Indeed, all current Ministerial mandate letters highlight the importance of interdepartmental cooperation and collaboration, particularly for science-based departments.
The success of the first GRDI shared priority projects (SPPs) has strongly demonstrated the feasibility and effectiveness of interdepartmental cooperation. These projects enable scientists in different departments to reach out to one another and combine their expertise to address issues that are beyond the mandates of single departments.
Shared Priority Projects
The first two large-scale shared priority research projects (2011-2016), the Quarantine and Invasive Species (QIS) project and the Food and Water Safety (FWS) project have performed beyond expectations—successfully demonstrating the ability of federal departments and agencies to work effectively together; leverage their research capacity and expertise; add value to existing resources; and build strong partnerships to deliver stronger results for Canadians than they ever could have managed on their own.
Recognizing both the unprecedented collaboration among departments as well as the outstanding results of the Food and Water Safety project and the Quarantine and Invasive Species project, the Government of Canada honoured the teams with a 2016 Public Service Award of Excellence for Scientific Contribution
Building on this success, two new shared priority projects are being conducted during Phase VI of the GRDI (2016-2021): the Antimicrobial Resistance project (AMR) and the Metagenomic-Based Ecosystem Biomonitoring project (EcoBiomics).
Food and Water Safety project (FWS)

Photo credit: Public Health Agency of Canada
More than 50 scientists from six federal departments and agencies were involved in the five-year project. The collaboration led to new, genomics-based technologies that detect, identify, and point to the potential source of pathogenic contaminants in food and water in hours rather than days, and with near-perfect accuracy. Teams from different departments collected and sequenced the DNA of hundreds of isolates of pathogens from farms, processing plants, grocery stores, lakes, rivers and other locations across Canada. These DNA sequences, along with a range of information about each of the isolates, have been added to a still-growing database.
The FWS project also provided the bioinformatics needed to manage and manipulate the data, developing a bioinformatics platform called IRIDA—Integrated Rapid Infectious Disease Analysis — now deployed across Canada, in collaboration with the BC Centre for Disease Control and Genome Canada-supported academics. Public health authorities have access to the capacity to not only quickly identify priority contaminants such as E. coli 0157:H7, Salmonella Enteritidis, listeria, and Campylobacter in a sample of food, but to compare its DNA signature to hundreds of samples of the bacteria collected across the country, providing them with valuable clues to the source of contamination.
This not only enables much faster and more effective responses to incidents of food or water contamination but, by providing a better understanding of the sources of contamination, will help to develop strategies to reduce the likelihood of these contaminants getting into our food and water supply in the first place.
Quarantine and Invasive Species project (QIS)
This five-year collaboration among some 29 research scientists representing a half-dozen departments and agencies developed genomics-based tools that have provided Canada with faster, more accurate and more cost-effective ways to identify a wide range of organisms, from freshwater finfish, aquatic parasites and insects to nematodes, fungi and plant viruses, among others.
In the past, it could take a week or more to determine whether an insect or microscopic pathogen detected at our border was a species that posed a threat—and even then, there was considerable margin for error, with potentially costly and irreversible consequences.
Leveraging for results
The GRDI involves extensive research relationships that extend across Canada and beyond. These additional collaborations include cooperative relationships with government-based science organizations, universities, industry, and other research institutes, both nationally and internationally, demonstrating the essential part the GRDI plays in leveraging the expertise and resources that help to accelerate progress in a cost-effective way. In the 2016-17 fiscal year, for example, GRDI projects attracted more than 480 collaborators external to the federal government:
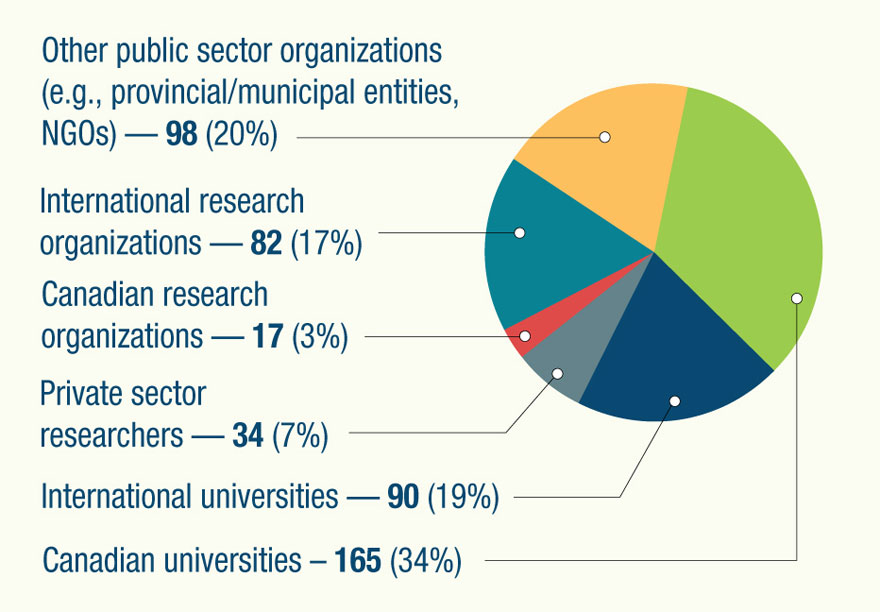
Long description of the "Leveraging for results"
- Canadian universities – 165 (34%)
- International universities — 90 (19%)
- International research organizations — 82 (17%)
- Canadian research organizations — 17 (3%)
- Private sector researchers — 34 (7%)
- Other public sector organizations (e.g., provincial/municipal entities, NGOs) — 98 (20%)
QIS researchers sequenced DNA from specimens collected from the environment as well as those held in government collections, building a database containing the genetic signature and other information about thousands of known organisms. The bioinformatics capacity developed as part of the project enables rapid comparison of DNA extracted from a new specimen against all the genetic signatures in the database in order to make a positive identification.
Antimicrobial Resistance project (AMR)
The development of resistance to antimicrobials by bacteria that were formerly sensitive is one of the most serious global health threats facing the world today. With no action to address microbial resistance, it is projected that by 2050, as many as 10 million deaths a year around the world will be attributable to bacterial infections that have developed resistance to antibiotics.
"... by 2050, as many as 10 million deaths a year around the world will be attributable to bacterial infections that have developed resistance to antibiotics."
The AMR project, involving scientists from five federal departments and agencies, uses a genomics-based approach to understand how food production contributes to the development of antimicrobial resistance of human health concern, and explore strategies for reducing antimicrobial resistance in food production systems.
The research focus is on food because of the widespread use of antibiotics in food production, and the impacts of this practice on the abundance of AMR bacteria in food products. Genomics insights into the impact of antibiotic alternatives on animal microbiomes will lead to new management practices that reduce antimicrobial use.
Protecting exports
Downey mildew of soybean is a quarantine species in very few countries. Still, Canada was challenged by Malaysia through the World Trade Organization to export soybeans that are free of the disease. This represents a fifty million dollar ($50M) annual market. Data assembled under the QIS project enabled scientists and their collaborators to rapidly answer specific questions regarding hosts and distribution, to help address this challenge. Malaysia changed its position on the basis of the evidence provided.
Data from the QIS project also allowed scientists to identify a Canadian species that causes a benign plant disease but that is the closest relative to a major corn disease, a US Select Agent, which could destroy 90% of all North American corn crops. Without this knowledge on the innocuous species, corn from Canada would likely test positive for the US select agent, potentially leading to major trade disruption.
The AMR project is a component of the Federal Action Plan on Antimicrobial Resistance and Use in Canada released in 2015, and will address public health, regulatory, and the agricultural sector's needs related to the use of antimicrobials. As an example of how ongoing GRDI research builds on previous accomplishments, AMR researchers are expanding the IRIDA bioinformatics platform developed by FWS project researchers to manage the molecular data they collect and "connect the dots" between AMR human clinical isolates and AMR bacteria in poultry, swine, beef and the environment.
Metagenomic-Based Ecosystem Biomonitoring project (EcoBiomics)
Biodiversity is paramount in water and soil to sustain diverse ecosystem services and economic activities across Canada. Land use disturbances are having enormous adverse impacts on the biodiversity and integrity of natural and managed ecosystems around the world. By the year 2100, the largest global impact on biodiversity is projected to be land use disturbance, followed by climate change, nutrient deposition, and new species introductions to ecosystems.
"... observatories will be located at scientifically important sites that are part of long-term monitoring programs in locations likely to be subject to increasing disturbances, such as the Great Lakes and in northern Alberta."
Land use disturbances impact soil biodiversity that sustains terrestrial ecosystems, and the healthy soils needed by the agricultural and forestry sectors in Canada. Disturbances such as excessive nutrient deposition impact the aquatic biodiversity that sustains aquatic ecosystems, and the clean water supplies needed for drinking, recreation, irrigation, and the fisheries sector in Canada.
A single gram of soil or 100 mL of river water can contain several million microorganisms, including bacteria, fungi, protozoa, and algae, and even more viruses. Genomics provides the only tools available to characterize this complex biodiversity. The Ecobiomics project will develop advanced genomics tools to assess freshwater ecosystem biodiversity and water quality in lakes and rivers; evaluate the health of soil essential to the productivity of agricultural and forestry systems across Canada; and investigate soil remediation for the oil and mining sectors. Genomics observatories will be located at scientifically important sites that are part of long-term monitoring programs in locations likely to be subject to increasing disturbances, such as the Great Lakes and in northern Alberta.
Addressing government priorities and departmental mandates
"... there is clear alignment of GRDI with the mandates and priorities of participating DAs [departments and agencies] and the federal government more generally. The evaluation confirmed that the program is consistent with, and contributes to, participating DAs' legislated mandates related to the health and safety of Canadians as well as the sustainability of Canada's natural resources (via regulatory activities) and support for industry (via economic development and regulatory activities). The evaluation further found that there is little duplication with other similar organizations or federal programs... In the absence of GRDI, the evaluation found that DAs' abilities to deliver on their mandates in the future might be negatively impacted as it is unclear what amount of DA funding for genomics R&D would be available given other DA priorities."
Innovation for jobs and prosperity
As described in the 2016 Goss Gilroy horizontal evaluation of the GRDI, the long-term benefits of GRDI-supported research are likely to be measured in the billions of dollars. Innovations based on GRDI research discoveries are poised to deliver social and economic benefits to Canadians across a wide range of sectors. Some examples:
Agriculture

Photo credit: Agriculture and Agri-Food Canada
The GRDI has funded research that promises to accelerate the development of varieties of wheat that are resistant to fusarium head blight—a disease that renders wheat unfit for consumption by humans or animals. The infection is spreading to ever-larger areas of major wheat-growing countries around the world, including Canada, where the disease has cost growers more than $1.5 billion since the mid-1990s. While some varieties of wheat show resistance to the disease, none are suitable for food production. They can be used as breeding stock, but it can take as long as 15 years to determine whether a new variety has potential as a commercial crop.
GRDI researchers have identified several specific elements of the wheat genome that appear to play a role in disease resistance. Combined with gene marker-assisted breeding techniques also developed through GRDI-funded research, the breeding process can be better targeted, and the commercial potential of a new variety determined in as little as five years.
Researchers funded through the GRDI have collaborated with the United States Department of Agriculture to accelerate the development of new and better varieties of oats. Canada is the world's second-biggest producer of oats, with more than $450 million a year in farm cash receipts. Given the proven health benefits of oats, growers are eager to find new varieties that offer even greater concentrations of heart-healthy fibre and antioxidants and that will perform well under a range of growing conditions. The development of new varieties has been slowed considerably by the size and complexity of the oat genome, which has made it difficult to apply the marker-assisted breeding technique used for other crops—oat breeders have had only a handful of genetic markers to work with.
That has changed dramatically. GRDI research has delivered thousands of gene markers based on their predictive value for traits such as grain yield, flowering time, disease resistance, and grain fiber concentration. These markers were used by oat breeders to produce superior oat varieties that were tested in the summer 2017 in replicated trials in Ottawa.
Fisheries

Photo credit: Fisheries and Oceans Canada
Research funded by the GRDI is helping to improve the management of Canada's multi-billion dollar seafood industry. For example, scientists used genomics to identify and map different populations of the Acadian Redfish and the Deepwater Redfish, species that are difficult to distinguish from one another. The research has enabled a greater understanding of Redfish stock structure and led to the development of separately managed Redfish fisheries where not previously possible. The research also played a key role in the designation of one population as 'threatened' by the Committee on the Status of Endangered Wildlife, leading to tailored conservation management strategies, and facilitating a partnership with Redfish harvesters to establish the first industry-led monitoring program for Redfish species—a clear demonstration of how genomics research improves conservation of at-risk stocks as well as utilization of healthy stocks for sustainable economic gains in Canada's coastal communities.
On the west coast, GRDI research will enable enhanced management of commercially and culturally important populations of Pacific Salmon. Hatcheries play a key part in Salmon stock management, releasing millions of juvenile Salmon every year to help maintain and rebuild vulnerable populations. Information obtained through tracking of hatchery fish contributes to the development of overall management strategies. Currently, hatchery fish are identified by wire tags implanted in juveniles prior to their release. Due to the cost involved—and the fact the tag cannot be retrieved without killing the fish—only about 10 percent of hatchery fish are tagged, limiting the information available to fishery managers.
GRDI researchers have developed tools that enable all broodstock in a hatchery to be genotyped—in essence, applying a genetic tag to all their offspring—meaning 100 percent of hatchery fish can be tagged for tracking. DNA samples can be retrieved non-invasively from fish caught in commercial or recreational fisheries, as well as those returning to spawn, enabling fish identification down to the individual parent to allow for better management and enhancement our economically and socially important Salmon stocks.
Forestry

Photo credit: Natural Resources Canada
Canada exports some $8 billion a year in softwood lumber alone—but access to international markets is not guaranteed. Countries reject imports of wood or wood products if any trace of a potentially damaging invasive species is found in a shipment.
These quarantine pests include the Pine Wood Nematode, or PWN, a microscopic worm that causes a disease that can kill a mature tree in a matter of weeks.
Even experts can have difficulty distinguishing the PWN from an extremely similar but harmless relative—creating the risk of a misidentification that can have significant economic consequences. GRDI-funded researchers have developed genomics-based tests that can accurately identify if unwanted pests, such as the PWN, are present in exported products. This test quickly identifies if the quarantine pest is present, giving Canadian exporters a cost-effective way to ensure their phytosanitary processes are effective in eliminating PWN from their wood products, and protecting Canada's export markets. This work received additional funding from the Canadian Food Inspection Agency, and has influenced international standard diagnostic protocols for PWN detection.
Among other resource management measures, forestry companies in Canada plant some 650 million trees every year—trees bred for strong growth, resistance to disease, adaptation to climate, and desirable wood characteristics. Unfortunately, breeding trees is a slow process—it can be 20 years or more before a tree reveals whether it has the hoped-for traits. GRDI funding is enabling researchers to study the genomes of mature trees, identifying genetic markers associated with the most sought-after qualities.
This creates the potential for breeders to select trees for further development at a very early stage—as seedlings or even as seeds—making the breeding process both more precise and significantly faster. The research has generated widespread interest, contributing to tree-breeding research being carried out by provincial governments in both Québec and Newfoundland and Labrador, and attracting funding and in-kind support from a range of public and academic institutions.
Environment
A GRDI-funded research project at Environment and Climate Change Canada (ECCC) has shown how genomics can enable precise identification of the causes of environmental problems. Even treated wastewater, for example, can contain thousands of bacteria, viruses, pharmaceuticals and industrial chemicals. Using traditional methods, it is very difficult to determine which of those may be responsible for a specific problem found downstream of the treatment outlet.
The researchers have demonstrated that, with toxicogenomics—the study of how an organism's genetic information responds to a toxin—it is possible to pinpoint exactly which toxins are causing the problem. Already, the city of Montreal has used the results of this research to support its decision to invest $250 million in an ozone-treatment system for wastewater. The same toxicogenomics techniques are being used in the Ecobiomics shared priority project to determine the extent to which oil sands development may be adding to natural toxicity from the bitumen that has been leaching into the ecosystem for thousands of years.
GRDI-funded research at ECCC is also transforming the way municipalities manage water safety at public beaches—often increasing effectiveness while reducing costs. For decades, communities have checked swimming areas for fecal bacteria contamination by testing for E. coli in the water. If the E. coli level was too high, the beach would be closed to swimming. It was usually assumed the fecal contamination was sewage overflow or pet droppings washed into the waterway after heavy rain. Using new genomics-based test methods, the ECCC researchers discovered that E. coli in water samples from beaches often matched the DNA fingerprint of E. coli from seagulls and Canada geese—not sewage or pet droppings. Now that the source of contamination can be identified with confidence, municipalities can better target mitigation measures. In Ottawa, for example, rather than investing in expensive sewage system upgrades, the city has installed overhead wires at some beaches to discourage gulls and geese. The research findings have generated interest from cities around the world.
Health
Research funded through the GRDI is helping to overcome a major hurdle on the way to realizing the huge potential of mesenchymal stem cells (MSCs) to treat a wide range of ailments. MSCs have a unique ability to help other types of stem cells repair damaged tissue but, like all stem cells, they also have the potential to mutate and become cancerous. And, just as they help other stem cells make repairs, they can also help cancerous stem cells grow more cancer. With a team of international collaborators, researchers at Health Canada have identified a series of protein and gene changes that indicate whether a particular MSC has a high probability of becoming cancerous. Based on these findings, the researchers are now focused on developing a rapid test platform that clinicians could use to screen MSCs to ensure their safety prior to use in treatment.
Salmonella is one of the most common causes of food-borne illness in Canada and around the world. While rarely fatal, the economic cost of Salmonella infection is measured in the billions of dollars a year. While rapid identification of the source of the bacteria is key to limiting the spread of illness, using traditional, animal-based test, it takes four days to identify the type of Salmonella found in a sample of food—information crucial to tracking the source of the contamination. In addition, only a handful of labs are equipped to perform the test.
A new, genomics-based test developed by GRDI researchers at the Public Health Agency of Canada, in collaboration with colleagues in the U.K. and Austria, reduces the time needed to type the bacteria to less than a day, enabling much faster response to incidents of contamination. Already accredited by the International Organization for Standardization, the new test can be done by almost any lab at minimal cost, so it's also ideal for routine surveillance for Salmonella at food processing plants. Based on this test, GRDI researchers have assembled computer algorithms to type and subtype Salmonella: the Salmonella in Silico Typing Resource—SISTR. Using this newly developed tool, the Canadian Food Inspection Agency and Canada's National Microbiology Laboratory are now able to quickly identify outbreaks of Salmonella and more effectively prevent the spread of this foodborne illness. SISTR has been made publicly available and researchers anywhere in the world can use this resource to type the Salmonella they want to identify. One such example is EnteroBase, an international centre for web-based bacterial genomic analysis at the University of Warwick in England which now uses this bioinformatics tool. Similarly, the U.S. Food and Drug Administration is evaluating SISTR as a potential tool in combatting foodborne illness. SISTR represents a significant contribution to reducing the public health risk posed by Salmonella not only in Canada, but around the world.
Gonorrhea is now a major public health concern as strains of the bacteria are increasingly showing resistance to currently used antibiotics. With the support of funding from the GRDI and working with researchers from the USA, UK, Australia, Sweden and the Netherlands, scientists at the Public Health Agency of Canada have created a sort of universal translator for drug resistance in gonorrhea—it's called NG-STAR, which stands for Neisseria gonorrhoeae Sequence Typing for Antimicrobial Resistance. Researchers around the world can refer to this database to identify the strain of gonorrhea they are working on. The Sanger Institute, one of the biggest and most influential centres for genomics research in the world, is working to incorporate NG-STAR into a broader gonorrhea sequencing project underway at Sanger headquarters at Oxford University in the UK.
Return on investment: a case study
In 2015, Health Canada asked Cambridge, Massachusetts-based Industrial Economics, Incorporated to evaluate the potential economic benefits of genomic techniques developed through GRDI-funded research to facilitate the tracking of the Campylobacter and Listeria bacteria.
While only rarely causing life-threatening or disabling illness, Campylobacter is responsible for some 130,000 cases of so-called food poisoning in Canada every year, resulting in significant costs to our health care system and additional costs in lost productivity. Although far less common—there are about 200 cases a year in Canada—Listeria infection is much more serious: in 2008, 24 people in Canada died as a result of an outbreak of listeriosis.
The evaluation concluded that the improved detection enabled by the GRDI-funded research could reduce the number of Campylobacter infections in Canada by at least 20 percent. Beyond the obvious health benefits, the study estimates this 20 percent reduction in the incidence of Campylobacter would lead to an annual economic benefit of more than $50 million. Similarly, the study predicted that genomics-based testing for Listeria would reduce the number of cases by a minimum 25 percent annually, reducing the cost to our economy by more than $70 million a year.
These minimum predicted economic benefits—more than $120 million a year—flow from a GRDI investment of less than $2.5 million.
The study further noted that, "By improving the ability of public health agencies to identify the source of specific strains of pathogen, it may also reduce the cost of unnecessary product recalls, or help to persuade industry of the need to adopt practices designed to reduce the risk of contamination that it might otherwise argue are unnecessary. If this proves to be the case, the benefits of the two research programs may prove to be even greater than those estimated here."
Source: Industrial Economics, Incorporated, Cambridge MA:
Economic Benefits of Genomics Research: Development of New Assays for Foodborne Pathogens (April 2015)
From discovery to application
From the beginning, the primary focus of the GRDI has been on achieving outcomes that provide tangible benefits to Canada and Canadians, and supporting the needs of end-users, whether in the public or private sectors. As demonstrated by the examples selected for inclusion in this summary report—many more of which are described on the GRDI website—GRDI-funded research is delivering a significant, positive impact for Canadians in a host of areas, and setting the stage for even greater benefits and further innovation in the future.
Given the range of the pioneering work undertaken through the GRDI, there is no single project or series of projects that could serve to exemplify the full extent of the impact of the initiative, and the clear link between GRDI discovery and application. Nonetheless, in this section of the report, two additional examples are provided, highlighting how research funded by the GRDI has earned Canada global recognition as a leader in the emerging field of toxicogenomics and for innovation in the application of genomics in cancer treatment:
Toxicogenomics
Research made possible by the GRDI has enabled Canadian researchers to place themselves among the leaders of a revolution in regulatory toxicology—the science behind the regulations that help to ensure everything from the medicines we take to the air we breathe is as safe as possible.
Traditionally, toxicity has been studied by dosing laboratory animals with a chemical—usually much more than would ever be encountered in daily life—and watching for the effects. It is a slow, expensive and somewhat inexact process. At Health Canada, a GRDI-funded project is showing how toxicogenomics can bring unrivalled precision and speed to regulatory toxicology, allowing researchers to see how different amounts of a chemical affect cells at the molecular level—and not only what the chemical does, but how it does it, all while reducing the need for animal testing.
These discoveries will allow scientists to provide regulators with more and better information about the toxicity of chemicals at less cost and in less time, leading to better regulation and greater protection for the health and safety of Canadians.
Their efforts have earned Canada a place on a prestigious international committee developing global strategies for toxicogenomics in regulatory policy—the benefits of which extend beyond prestige. As a researcher at Health Canada has pointed out, "To have a say and influence in the development of international guidelines—rather than having to adapt to the guidelines or standards set by others—is a real advantage."
Cancer treatment
Over several phases of GRDI support, the National Research Council of Canada (NRC) has built an impressive target discovery and antibody development pipeline, primarily for oncology indications. Using a combination of genomic, proteomic and bioinformatic approaches, promising targets are identified based on their cancer-associated profiles. Hundreds of antibodies are then made against these targets and screened for specificity and function.

Photo credit: Agriculture and Agri-Food Canada
Of the traditional antibodies discovered by NRC through this pipeline, one licensed to Montreal-based Alethia Biotherapeutics is the most advanced. In May 2015, the company announced the beginning of patient dosing in a Phase 1 (first in human) clinical trial for its fully humanized monoclonal antibody against cancer, co-developed with NRC with support from the GRDI.
NRC scientists are now adapting this pipeline to the latest immunotherapy modality: antibody-drug conjugates, or ADCs—a type of biotherapeutic that uses an antibody with a drug attached to it to target and destroy cancer cells, with minimal impact on surrounding tissue. A collaboration with a private sector pharmaceutical company has led to the development of an ADC that has proven successful in shrinking tumours in preclinical models and is now approved for Phase 1 trials in humans.
By working with the different actors of the Canadian biopharmaceutical ecosystem, including contract research and manufacturing organizations, NRC enables the advancement of this next generation of biotherapeutics. These interactions are stimulating the growth of the sector thereby creating jobs and increasing Canada's return on its innovation investments.
Conclusion: Efficient, Effective, Necessary
As noted at the beginning, this report can provide only a sampling of the positive impacts of GRDI-funded research and how the emphasis on collaboration magnifies the Government of Canada's investment in the program. The 2016 Goss Gilroy evaluation of the GRDI provides an excellent summary of the overall impacts of the GRDI:
"In spite of large amounts of funds being leveraged and some DAs [departments and agencies] making investments in genomics R&D, it is clear that the need for continued support for these types of projects does not show signs of abating. In the absence of GRDI, the evaluation found that DAs' abilities to deliver on their mandates in the future might be negatively impacted as it is unclear what amount of DA funding for genomics R&D would be available given other DA priorities.
The continued need for GRDI is taking place within a context of a decreasing real value of the funding (due to inflation since inception in 1999 without commensurate increases in fundingFootnote 3), funding being shared among more participating DAs, SPPs vying for part of the GRDI funding envelope and increasing costs associated with the research. Research costs are increasing because more effort is now being focused on applications and transferring knowledge to end-users, which are more resource intensive phases of R&D.
In terms of effectiveness, the evaluation found that Phase V projects have been successful in the development of innovative knowledge and technologies and influencing evidence-based public policy.
The program has produced expected outputs and exceeded targets related to these. Moreover, the evaluation confirmed that many GRDI projects have successfully transferred knowledge and technologies to end-users, both internal and external to the federal government. Many more GRDI projects have the potential to similarly impact end-users.
In terms of efficiency and effectiveness, the evaluation found that the program is both efficient (in terms of its horizontal governance) and effective (in terms of its use of collaborations, leveraging of funds and delivery approach). The evaluation did not reveal any significant opportunities for improvement in program governance or delivery."Footnote 4